
- #SERIAL CLONER FEATURE IMPORT GENBANK GENERATOR#
- #SERIAL CLONER FEATURE IMPORT GENBANK SERIAL#
- #SERIAL CLONER FEATURE IMPORT GENBANK SOFTWARE#
Cavanaugh, Mark Clark, Karen Karsch-Mizrachi, Ilene Lipman, David J. Sucrose synthase gene sequences of various cereals like rice, maize, and barley were accessed from NCBI Genbank database.īenson, Dennis A. sequencing of sucrose synthase gene fragment from sor- ghum using primers designed at their conserved exons. Sequence analysis of cereal sucrose synthase genes and isolation. GenBank is designed to provide and encourage access. Department of Health & Human Services — GenBank is the NIH genetic sequence database, an annotated collection of all publicly available DNA sequences. Data exchange with the EMBL Data Library and the DNA Data Bank of Japan helps ensure comprehensive worldwide coverage. Most submissions are made using the BankIt (web) or Sequin programs and accession numbers are assigned by GenBank staff upon receipt. The GenBank sequence database incorporates publicly available DNA sequences of more than 105 000 different organisms, primarily through direct submission of sequence data from individual laboratories and large-scale sequencing projects. It is also possible to copy sequences in a formatted or unformatted way in the clipboard.Benson, Dennis A.

It is also possible to save (or copy in the clipboard) multiple sequences to submit a multi-alignement request to Clustal.
#SERIAL CLONER FEATURE IMPORT GENBANK SERIAL#
Serial Cloner also allows to export GenBank files as well as Fasta-formatted text files.
#SERIAL CLONER FEATURE IMPORT GENBANK GENERATOR#
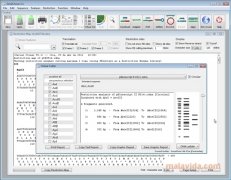
A virtual cutter, a web browser with instant import of NCBI/EMBL entries and a silent restriction map generator are also provided. Features of the aligned sequences are displayed. During all these sub-cloning steps, Features present in plasmids and insert will be properly inherited.įinally, Serial Cloner provides an interface to align two sequences using a local algorithm or the BLAST2Seq NCBI server and extract a consensus. An additional interface allows easy Gateway(tm) cloning for both BP and LR reactions. Just select, blunt if you need, and click the Ligate button. You can assemble fragments, obtained by PCR, adaptor/shRNA synthesis or simply by graphically selecting fragments between restriction sites. shRNA constructions based on pre-defined scaffolds are also automated. In addition to the classical restriction maps - both graphic and text-based - or site usage windows, you will be able to quickly set-up a PCR-based amplification or prepare synthetic adaptors. Serial Cloner will assist you in setting-up new sub-cloning projects and in preparing the electronic versions of your constructs. Features can be added to protein sequences and inherited from DNA upon translation (done using chosen genetic code). Serial Cloner handles Annotations and Features both in the sequence and in the Graphic Map and can automatically scan for sequence Features. Text-based Sequence maps are also generated and also display Features. The graphic map of Serial Cloner is really Graphic as you can easily select and extract a feature, a fragment or show single, double or multiple cutter all in the same window. Serial Cloner also lets you build text restriction map and quickly format it to add multi-frame translation or only show single cutters for example. All key functions are accessible through a compact interface. All the tools you need to analyze and manipulate your sequences are available in an all-in-one-window concept. Powerful graphical display tools and simple interfaces help the analysis and construction steps in a very intuitive way. Version 2.6 can now estimate the isolectric point of proteins using various methods. Serial Cloner can open protein sequences, align and reverse-translate them using defined codon-biais tables. The included Web interface also allows to submit BLAST request directly from Serial Cloner Sequence windows. Genbank/EMBL files can also be read or imported from internet using the included web interface. Serial Cloner can read and write DNA Strider-compatible files and also import or export files in FASTA format, MacVector, VectorNTI, ApE and GenBank files with their features as well as pDRAW32 sequences. User can annotate any sequence portion or use the automatic scan to detect common features. Serial Cloner 2.6 handles Features/Annotations and allows to define, import and export, customized lists of Features.
#SERIAL CLONER FEATURE IMPORT GENBANK SOFTWARE#
Serial Cloner is a Molecular Biology software that provides tools with an intuitive interface to assists you in DNA cloning, sequence analysis and visualization.


 0 kommentar(er)
0 kommentar(er)
